Research
>>>Yue Sun, Limei Wang, Kun Gao, Shihui Ying, Weili Lin, Kathryn L. Humphreys, Gang Li, Sijie Niu, Mingxia Liu*, Li Wang∗, “Self-Supervised Learning with Application for Infant Cerebellum Segmentation and Analysis”, in Nature Communications, vol. 14, 4717, 2023.
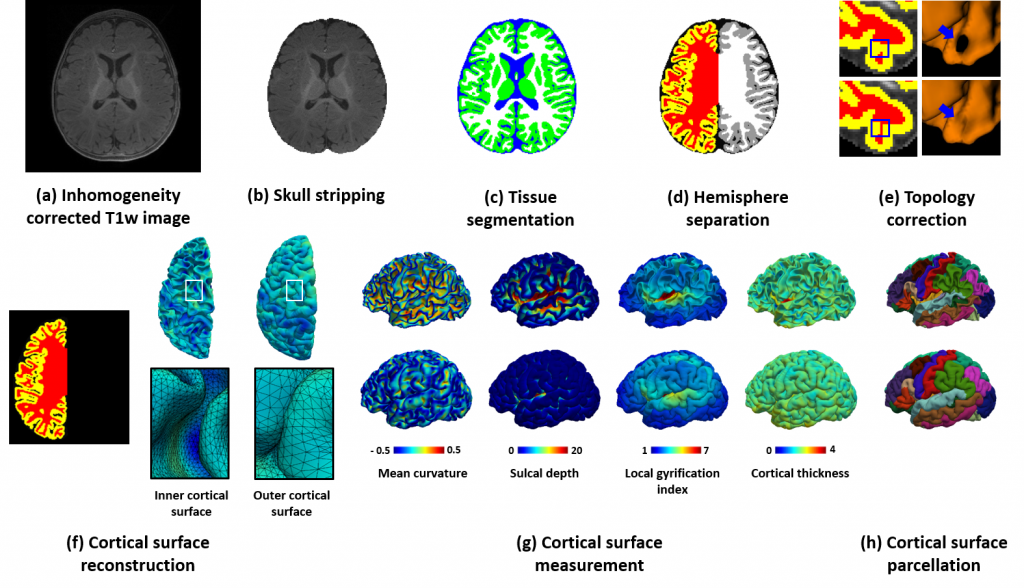
>>> Li Wang∗,#, Zhengwang Wu∗,#, Liangjun Chen, Yue Sun, Weili Lin, Gang Li∗, “iBEAT V2.0: A Multi-site Applicable, Deep Learning-based Pipeline for Infant Cerebral Cortical Surface Reconstruction”, in Nature Protocols, vol. 18, no. 5, pp. 1488–1509, 2023.
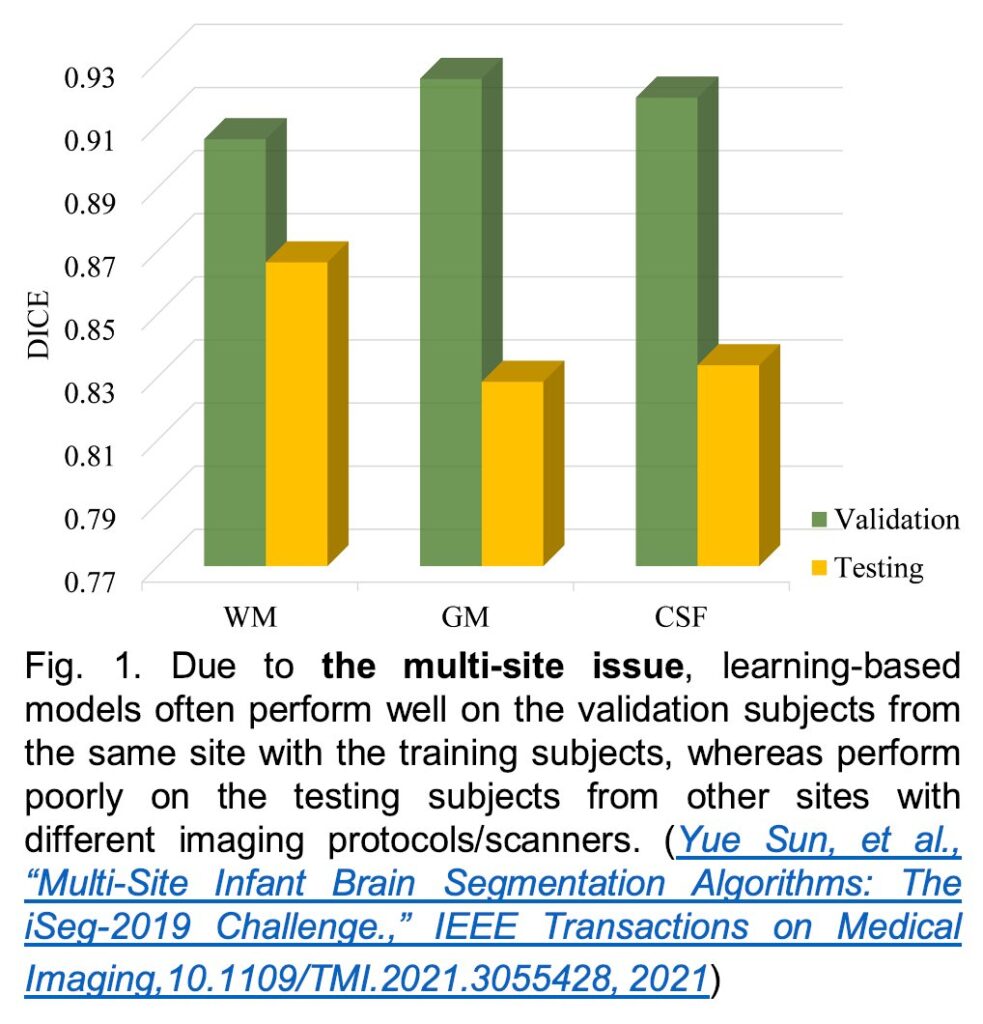
>>> Multi-Site Infant Brain Segmentation Algorithms: The iSeg-2019 Challenge, IEEE Transactions on Medical Imaging, 40(5), 1363-1376, 2021. (Download for personal use) Appendix
To better understand early brain development in health and disorder, it is critical to accurately segment infant brain magnetic resonance (MR) images into white matter (WM), gray matter (GM), and cerebrospinal fluid (CSF). Deep learning-based methods have achieved state-of-the-art performance; however, one of the major limitations is that the learning-based methods may suffer from the multi-site issue, that is, the models trained on a dataset from one site may not be applicable to the datasets acquired from other sites with different imaging protocols/scanners. To promote methodological development in the community, the iSeg-2019 challenge (http://iseg2019.web.unc.edu) provides a set of 6-month infant subjects from multiple sites with different protocols/scanners for the participating methods. Training/validation subjects are from UNC (MAP) and testing subjects are from UNC/UMN (BCP), Stanford University, and Emory University. By the time of writing, there are 30 automatic segmentation methods participated in the iSeg-2019. In this article, 8 top-ranked methods were reviewed by detailing their pipelines/implementations, presenting experimental results, and evaluating performance across different sites in terms of whole brain, regions of interest, and gyral landmark curves. We further pointed out their limitations and possible directions for addressing the multi-site issue. We find that multi-site consistency is still an open issue. We hope that the multi-site dataset in the iSeg-2019 and this review article will attract more researchers to address the challenging and critical multi-site issue in practice.
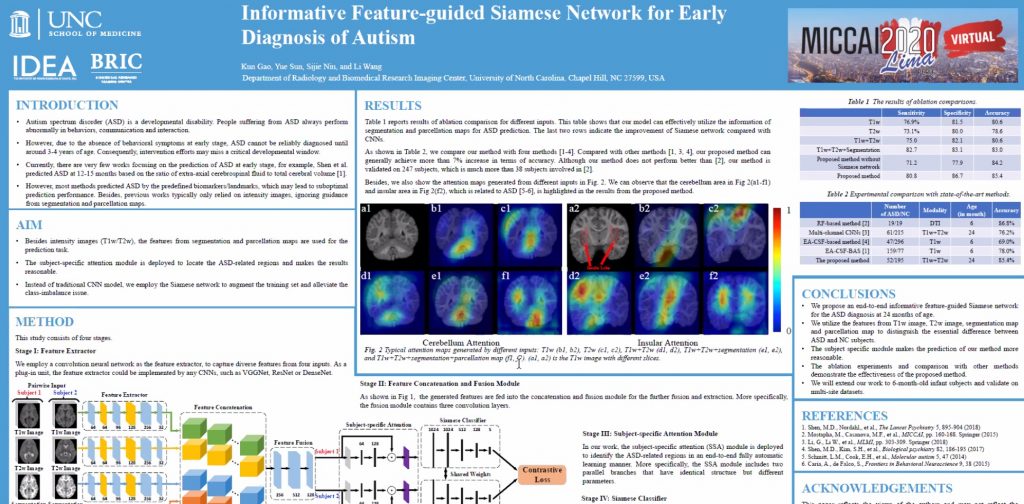
>>> Informative Feature-guided Siamese Network for Early Diagnosis of ASD – presented in MLMI 2020: PDF Talk
Autism, or autism spectrum disorder (ASD), is a complex developmental disability, and usually diagnosed with observations at around 3-4 years old based on behaviors. Studies have indicated that early treatment, especially during early brain development in the first two years of life, can significantly improve the symptoms, therefore, it is important to identify ASD as early as possible. Most previous works employed imaging-based biomarkers for the early diagnosis of ASD. However, they only focused on extracting features from the intensity images, ignoring the more informative guidance from segmentation and parcellation maps. Moreover, since the number of autistic subjects is always much smaller than that of normal subjects, this class-imbalance issue makes the ASD diagnosis more challenging. In this work, we propose an end-to-end informative feature-guided Siamese network for the early ASD diagnosis. Specifically, besides T1w and T2w images, the discriminative features from segmentation and parcellation maps are also employed to train the model. To alleviate the class-imbalance issue, the Siamese network is utilized to effectively learn what makes the pair of inputs belong to the same class or different classes. Furthermore, the subject-specific attention module is incorporated to identify the ASD-related regions in an end-to-end fully automatic learning manner. Both ablation study and comparisons demonstrate the effectiveness of the proposed method, achieving an overall accuracy of 85.4%, sensitivity of 80.8%, and specificity of 86.7%.
Kun Gao, Yue Sun, Sijie Niu, and Li Wang Department of Radiology and Biomedical Research Imaging Center, University of North Carolina at Chapel Hill, USA li_wang@med.unc.edu
>>>Semi-supervised Transfer Learning for Infant Cerebellum Tissue Segmentation – presented in MLMI 2020 PDF TALK
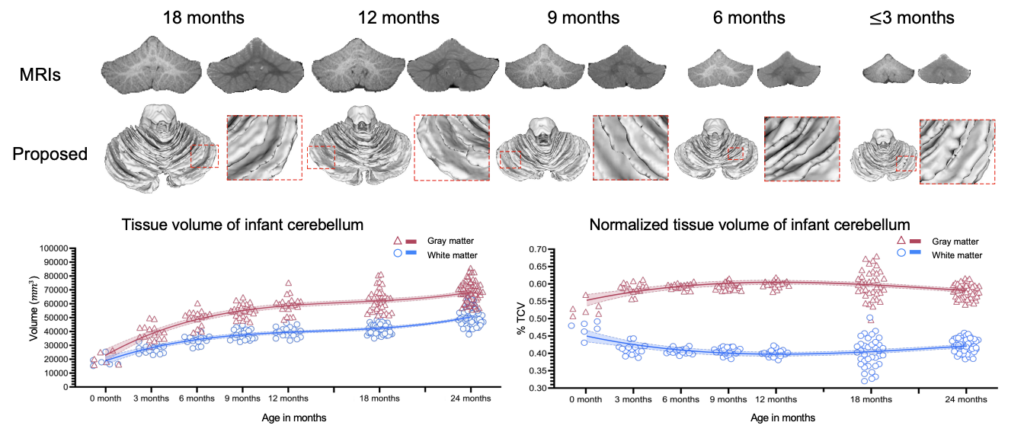
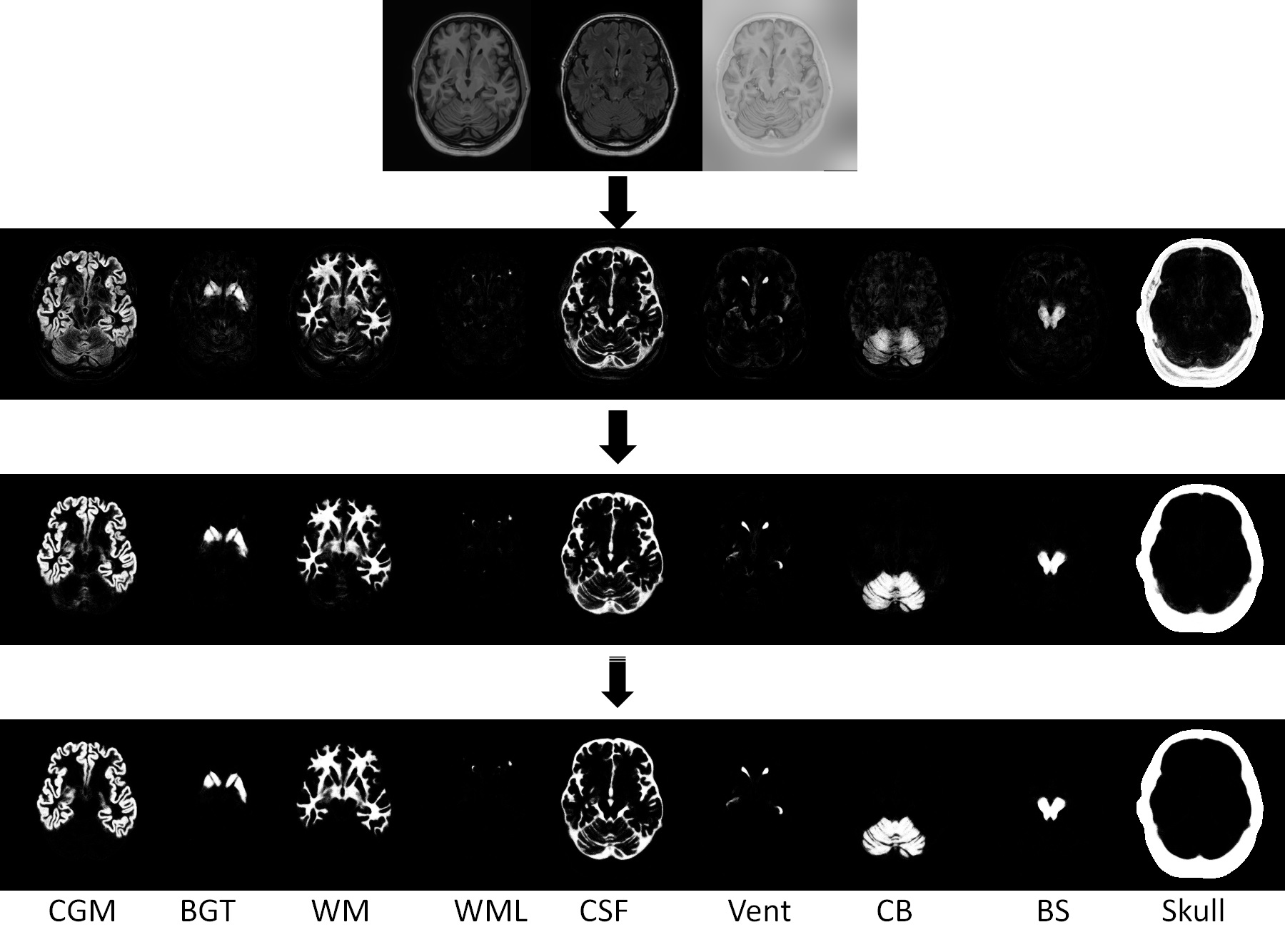
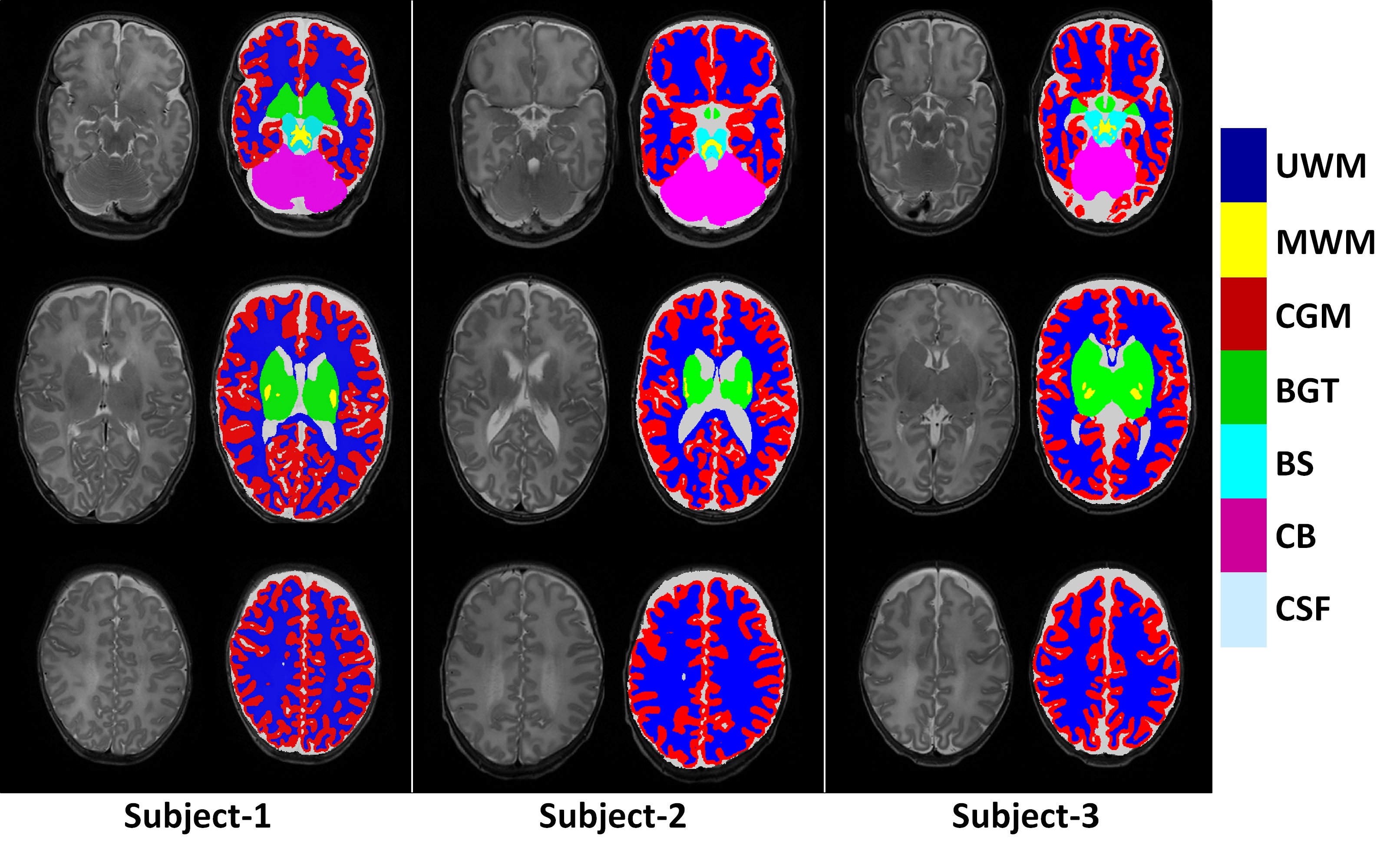
To characterize early cerebellum development, accurate segmentation of the cerebellum into white matter (WM), gray matter (GM), and cerebrospinal fluid (CSF) tissues is one of the most pivotal steps. However, due to the weak tissue contrast, extremely folded tiny structures, and severe partial volume effect, infant cerebellum tissue segmentation is especially challenging, and the manual labels are hard to obtain and correct for learning-based methods. To the best of our knowledge, there is no work on the cerebellum segmentation for infant subjects less than 24 months of age. In this work, we develop a semi-supervised transfer learning framework guided by a confidence map for tissue segmentation of cerebellum MR images from 24-month-old to 6-month-old infants. Note that only 24-month-old subjects have reliable manual labels for training, due to their high tissue contrast. Through the proposed semi-supervised transfer learning, the labels from 24-month-old subjects are gradually propagated to the 18-, 12-, and 6-month-old subjects, which have a low tissue contrast. Comparison with the state-of-the-art methods demonstrates the superior performance of the proposed method, especially for 6-month-old subjects.
Yue Sun, Kun Gao, Sijie Niu, Weili Lin, Gang Li, Li Wang, and the UNC/UMN Baby Connectome Project Consortium Department of Radiology and Biomedical Research Imaging Center, University of North Carolina at Chapel Hill, USA li_wang@med.unc.edu
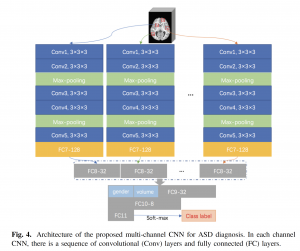
>>> Early Diagnosis of Autism Disease by Multi-Channel CNNs
Li, Guannan, Mingxia Liu, Quansen Sun, Dinggang Shen, and Li Wang. “Early Diagnosis of Autism Disease by Multi-channel CNNs.” In International Workshop on Machine Learning in Medical Imaging, pp. 303-309. Springer, Cham, 2018.
Currently there are still no early biomarkers to detect infants with risk of autism spectrum disorder (ASD), which is mainly diagnosed based on behavior observations at three or four years old. Since intervention efforts may miss a critical developmental window after 2 years old, it is significant to identify imaging-based biomarkers for early diagnosis of ASD. Although some methods using magnetic resonance imaging (MRI) for brain disease prediction have been proposed in the last decade, few of them were developed for predicting ASD in early age. Inspired by deep multi-instance learning, in this paper, we propose a patch-level data-expanding strategy for multi-channel convolutional neural networks to automatically identify infants with risk of ASD in early age. Experiments were conducted on the National Database for Autism Research (NDAR), with results showing that our proposed method can significantly improve the performance of early diagnosis of ASD.
>>> Volume-based Analysis of 6-month-old Infant Brain MRI for Autism BioMarker Identification and Early Diagnosis [PDF] [Code: Caffe prototxt] [Software]
Autism spectrum disorder (ASD) is mainly diagnosed by the observation of core behavioral symptoms. Due to the absence of early biomarkers to detect infants either with or at–risk of ASD during the first postnatal year of life, diagnosis must rely on behavioral observations long after birth. As a result, the window of opportunity for effective intervention may have passed when the disorder is detected. Therefore, it is clinically urgent to identify imaging-based biomarkers for early diagnosis and intervention. In this paper, for the first time, we proposed a volume-based analysis of infant subjects with risk of ASD at very early age, i.e., as early as at 6 months of age. A critical part of volume-based analysis is to accurately segment 6-month-old infant brain MRI scans into different regions of interest, e.g., white matter, gray matter, and cerebrospinal fluid. This is actually very challenging since the tissue contrast at 6-month-old is extremely low, caused by inherent ongoing myelination and maturation. To address this challenge, we propose an anatomy-guided, densely-connected network for accurate tissue segmentation. Based on tissue segmentations, we further perform brain parcellation and statistical analysis to identify those significantly different regions between autistic and normal subjects. Experimental results on National Database for Autism Research (NDAR) show the advantages of our proposed method in terms of both segmentation accuracy and diagnosis accuracy over state-of-the-art results.
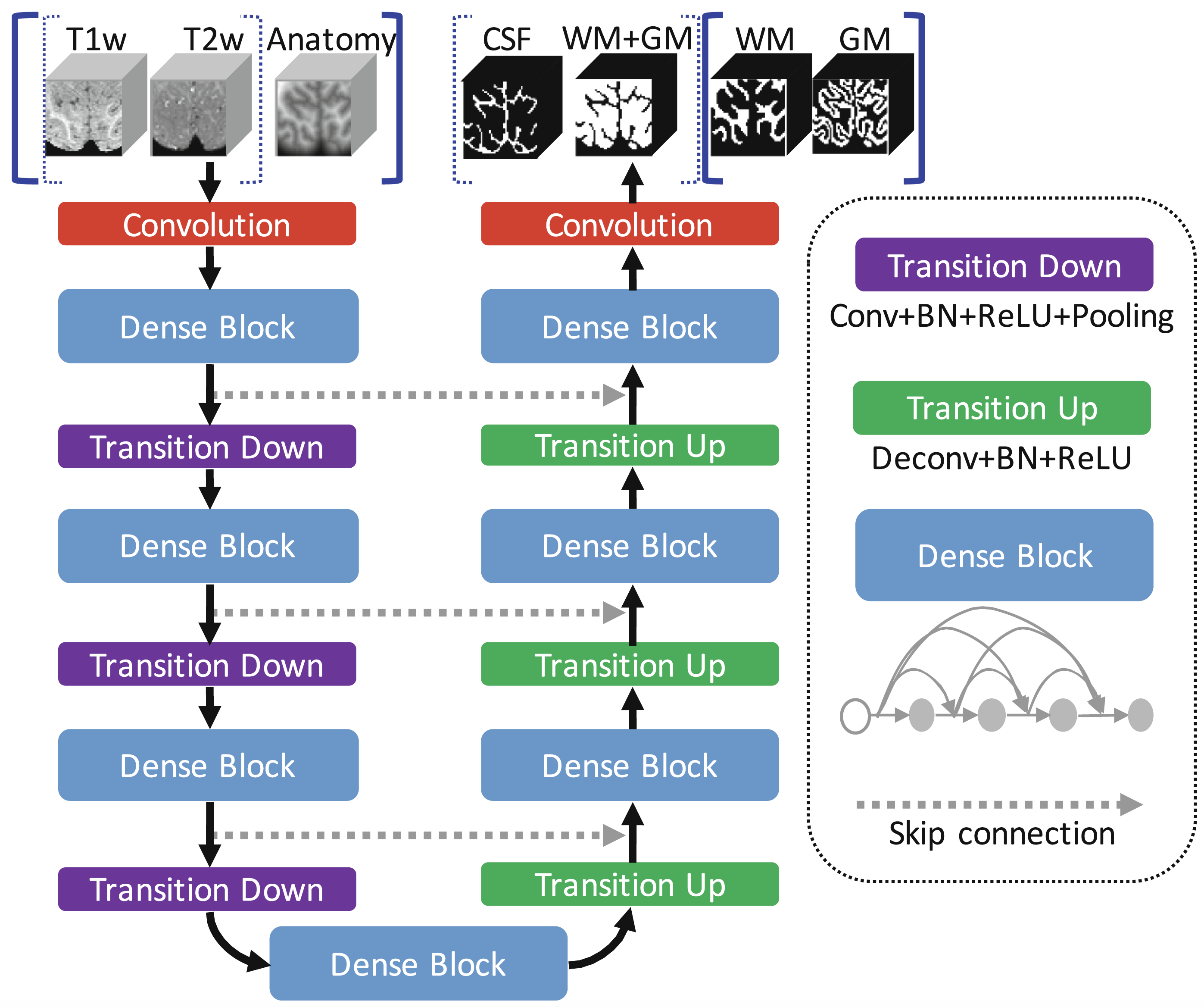
Anatomy-Guided Densely-Connected U-Net:
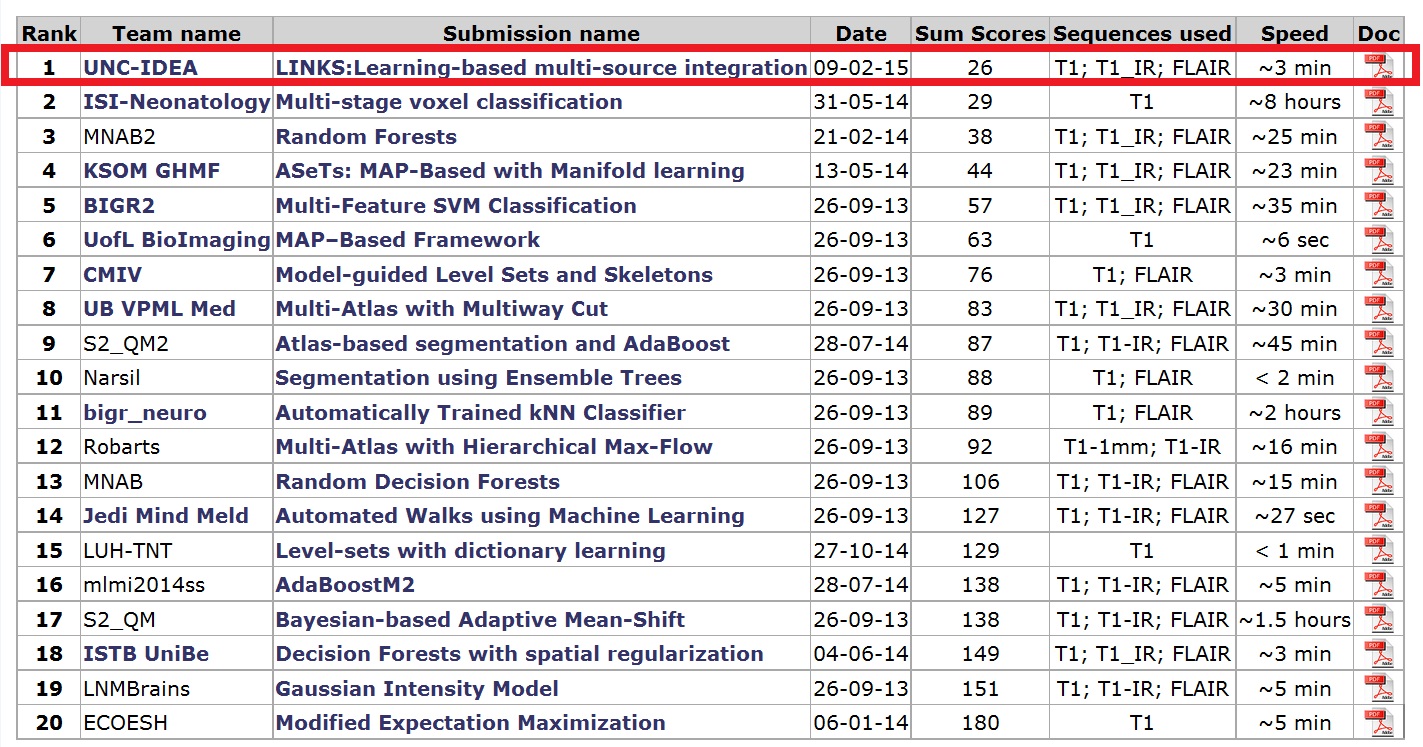
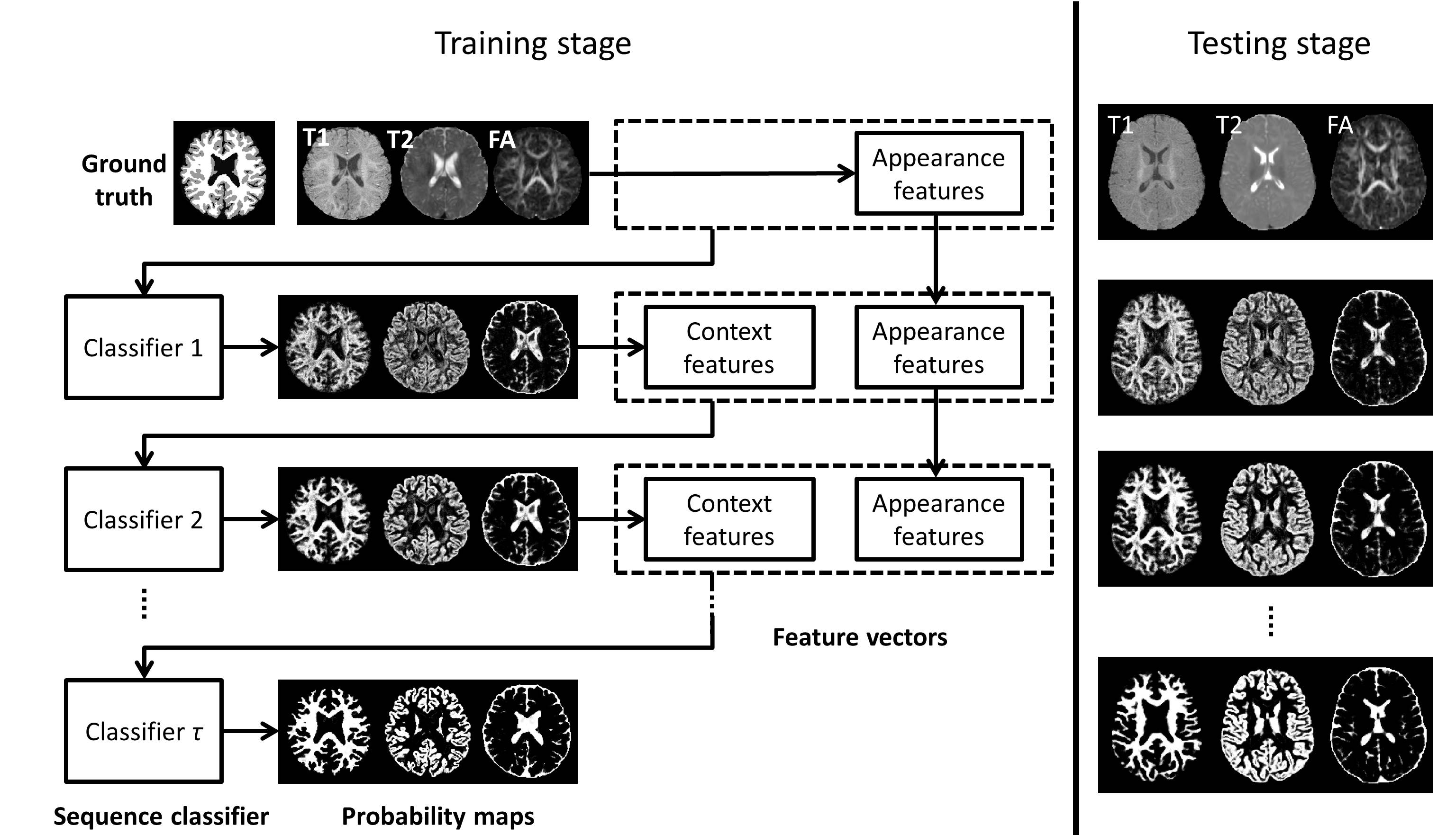
>>> LINKS: Learning-based multi-source IntegratioN frameworK for Segmentation of infant brain images. This novel method employs the random forest and auto-context model. [Journal version] [PPT] [Email request for the code]
The proposed method can be applied on brain images at any time phase of lifespan, from 2-weeks neonatal brain to elderly brain. Especially, our method has achieved #1 rank on both neonatal (< 3 months of age) brain segmentation MICCAI challenge and MR brain (> 12 months of age) segmentation MICCAI challenge.
The following shows the LINKS framework for the training stage and application stage on the isointense (~6 months old of age) infant brain images. Such isointense infant brain images have an extramely low tissue contrast due to myelination, which results in a very challengeing task for the segmentation:
We have applied this framework on the MRBrainS13 MICCAI Challenge data. The following shows the segmentation results on a target brain image by our method. Our model achieves the #1st place. For more info, please refer to the MICCAI MRBrainS13 Challenge.
We also have applied this framework on the NeoBrainS12 MICCAI Challenge data. The following shows the segmentation results on 3 target neonatal images by our method. Our model achieves the #1st place. For more info, please refer to the NeoBrainS12 Challenge.
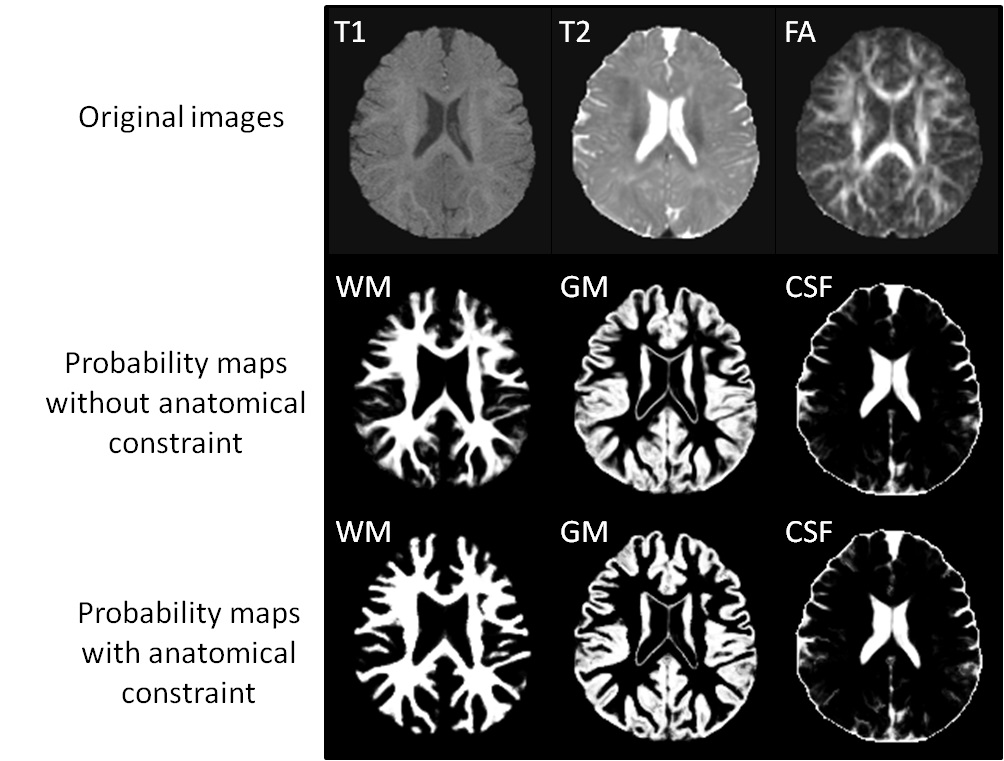
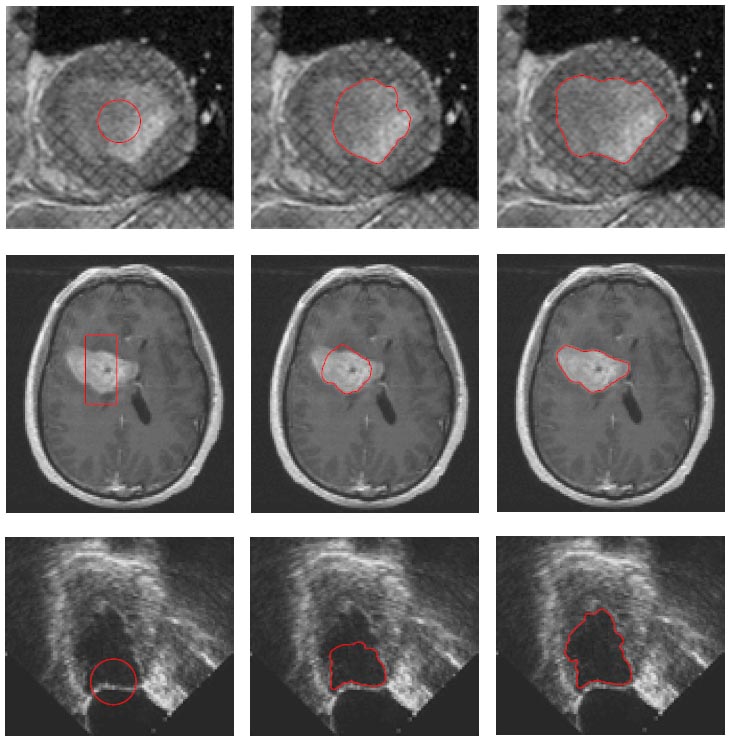
>>> Integration of Sparse Multi-modality Representation and Geometrical Constraint for Isointense Infant Brain Segmentation. [PDF][Code will be released soon]
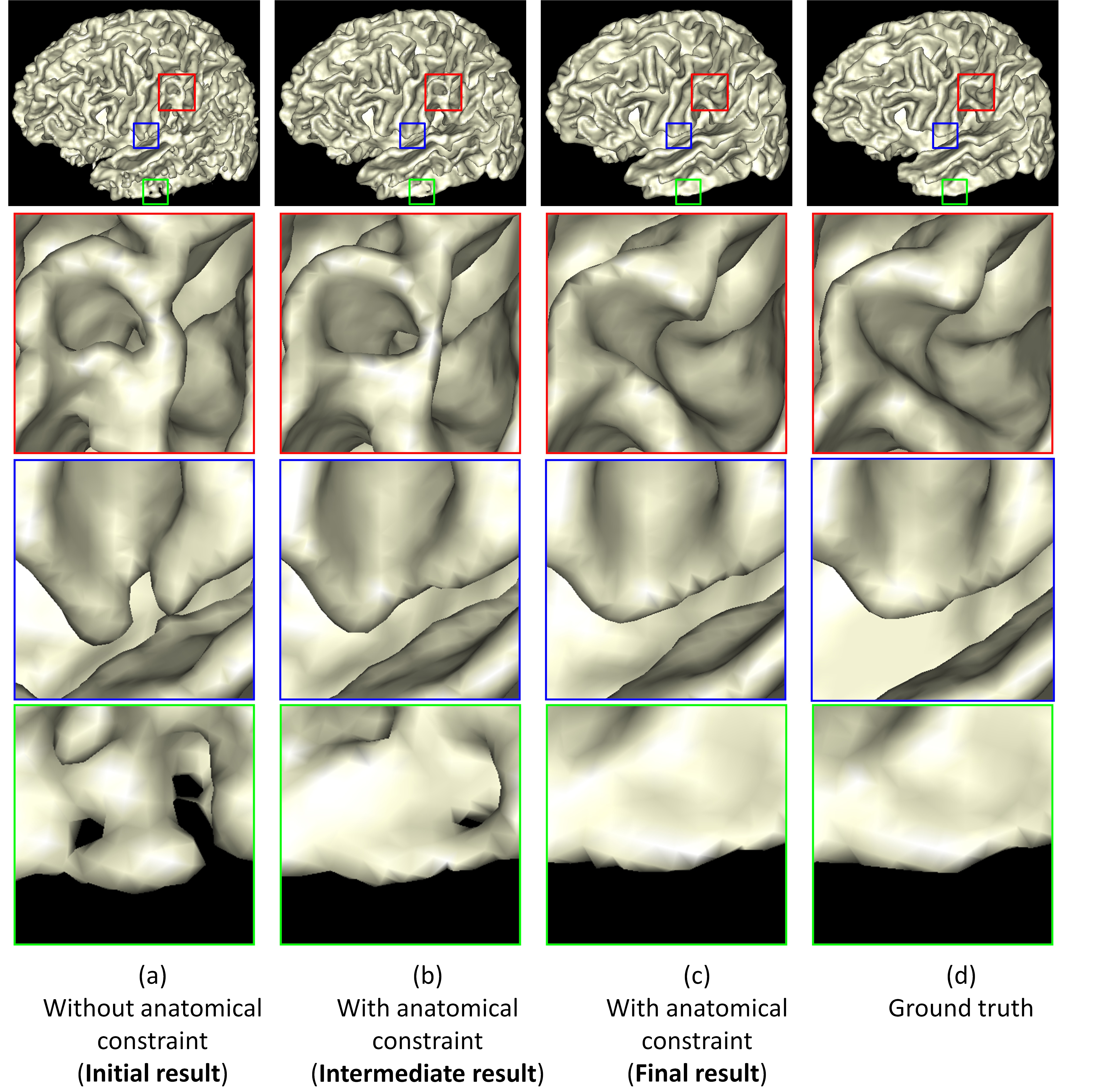
>>>>> Neonatal Brain MR Image Segmentation using Sparse Representation and Patch-Driven Level Sets. [PDF][PPT][Matlab Source Code]
>>> Longitudinally guided level sets for consistent tissue segmentation of neonates. [PDF] [Infant processing software]
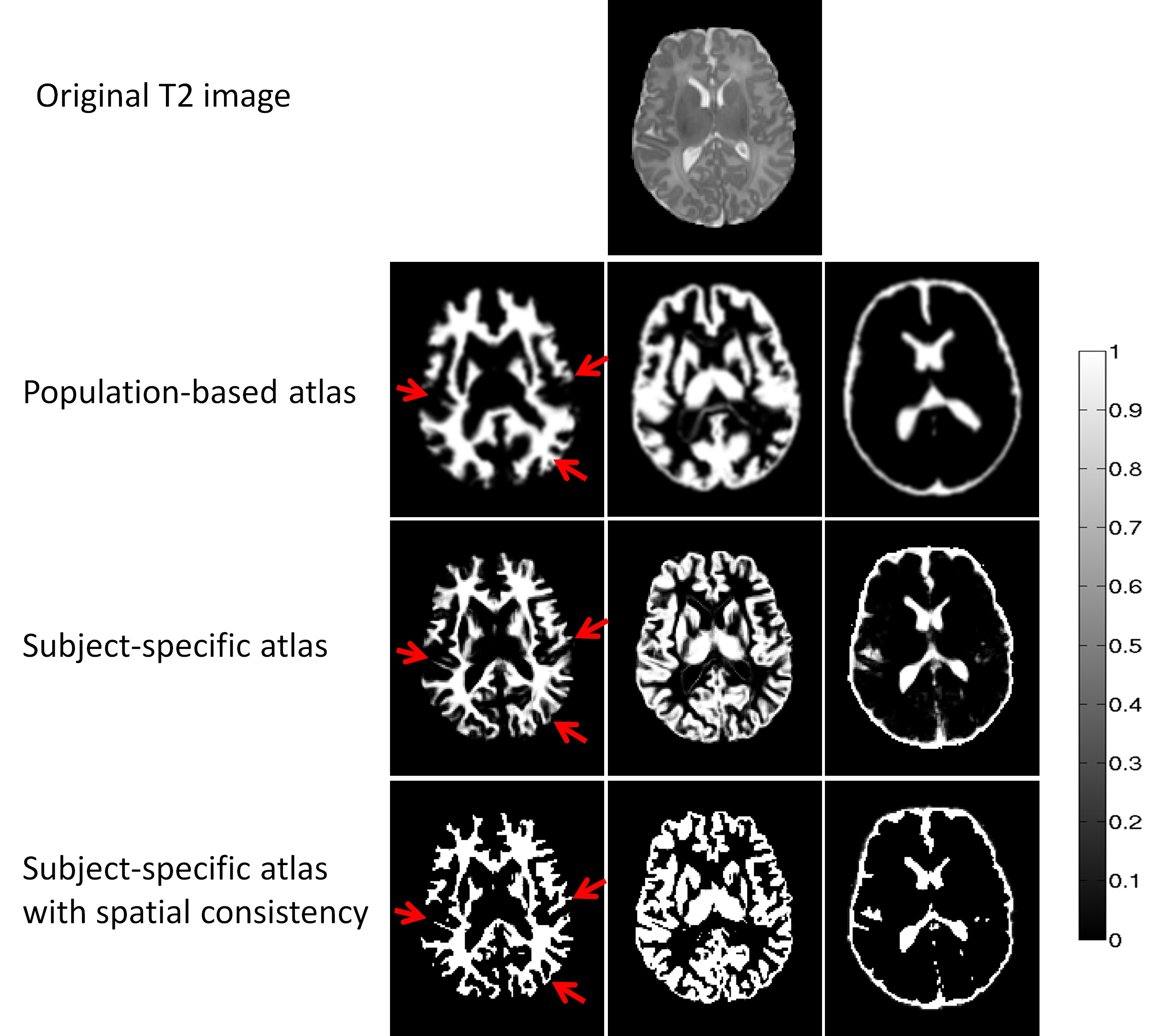
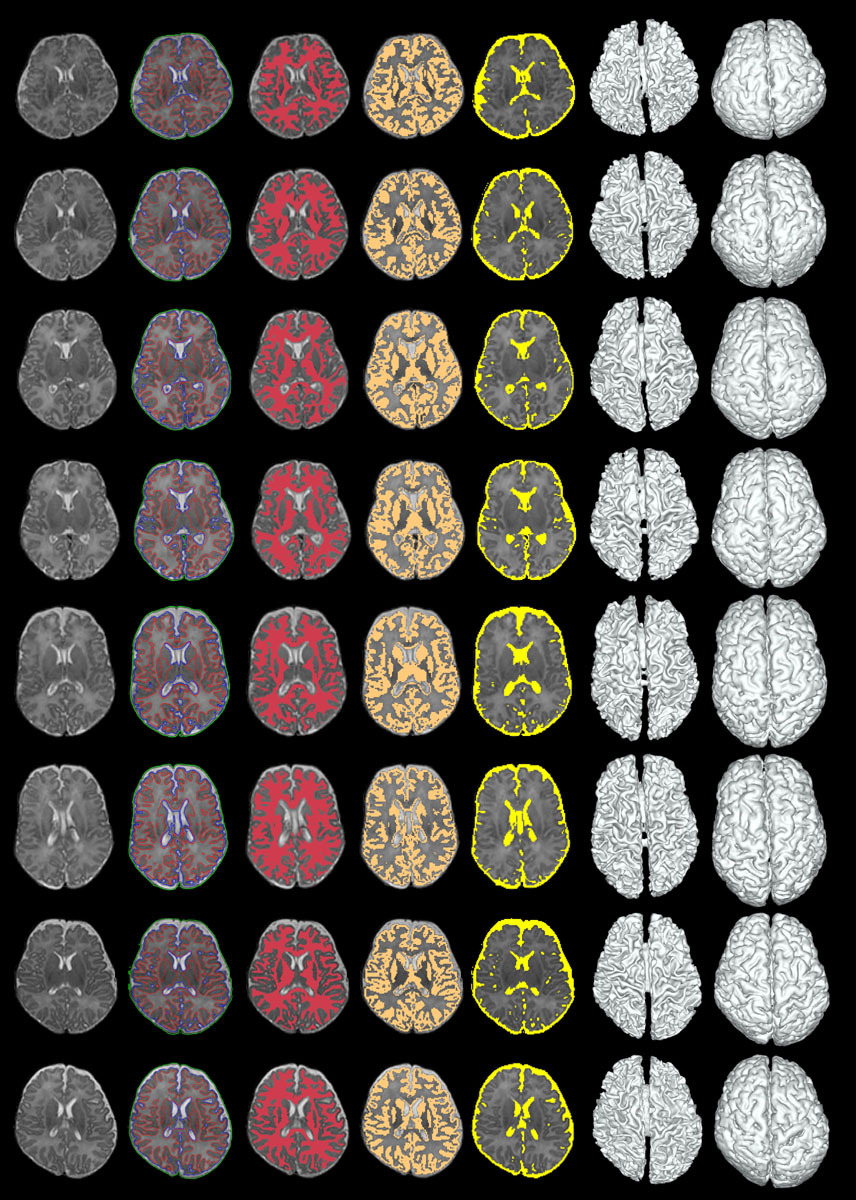
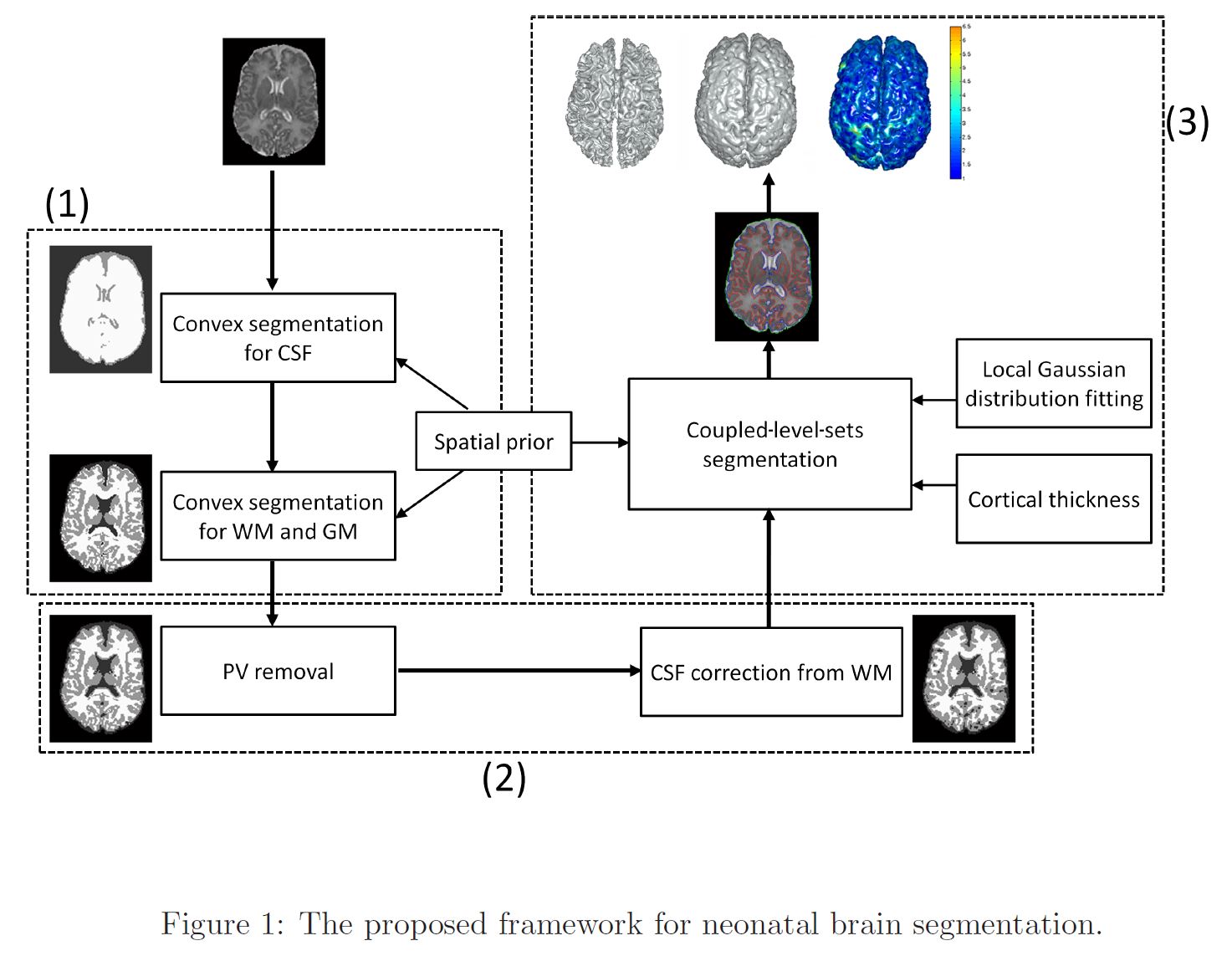
>>> Automatic Segmentation of Neonatal Images Using Convex Optimization and Coupled Level Sets. [PDF][Infant processing software]
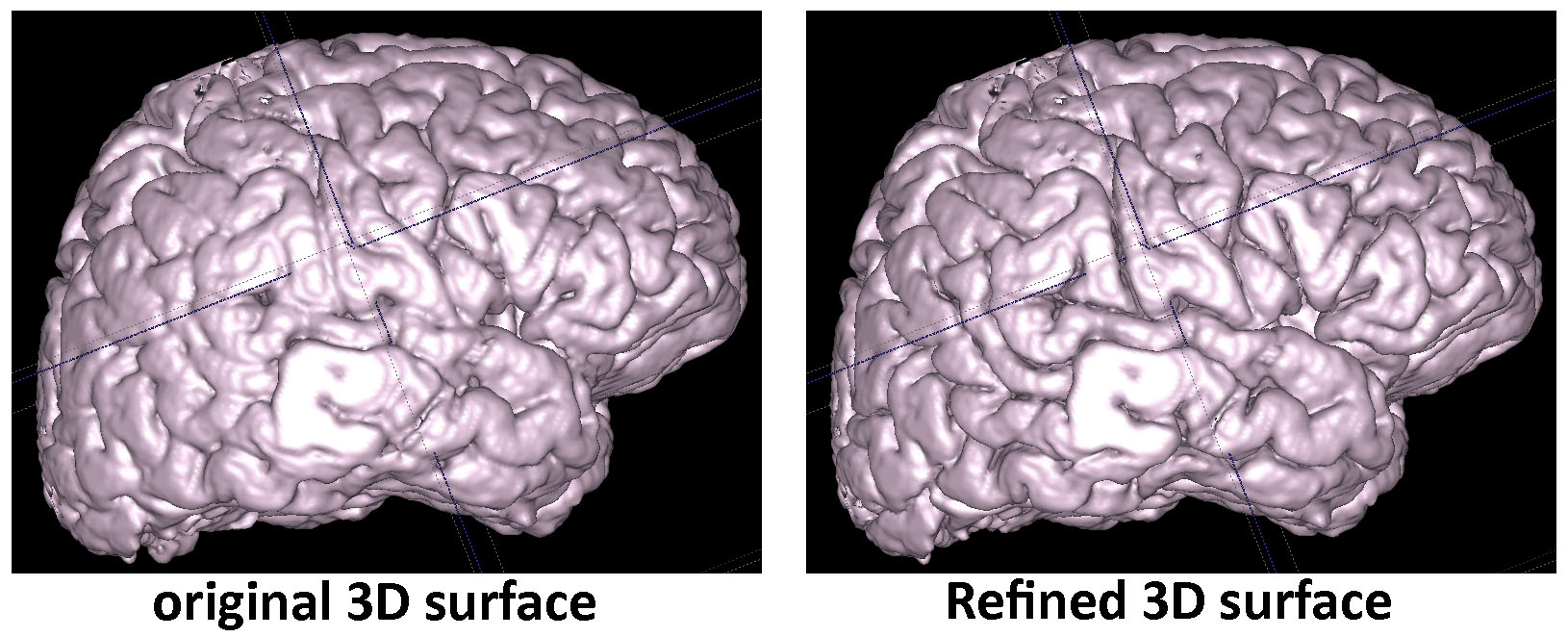
>>> Extraction of the Cerebral Cortical Boundaries (TBA) (click the image to see the high resolution)
>>> Medical Image Segmentation with Local Gaussian Distribution (LGD) Fitting Energy [PDF][Matlab Source Code]